A Novel Method for Characterization of Biotic-Abiotic Interactions
December 03, 2020
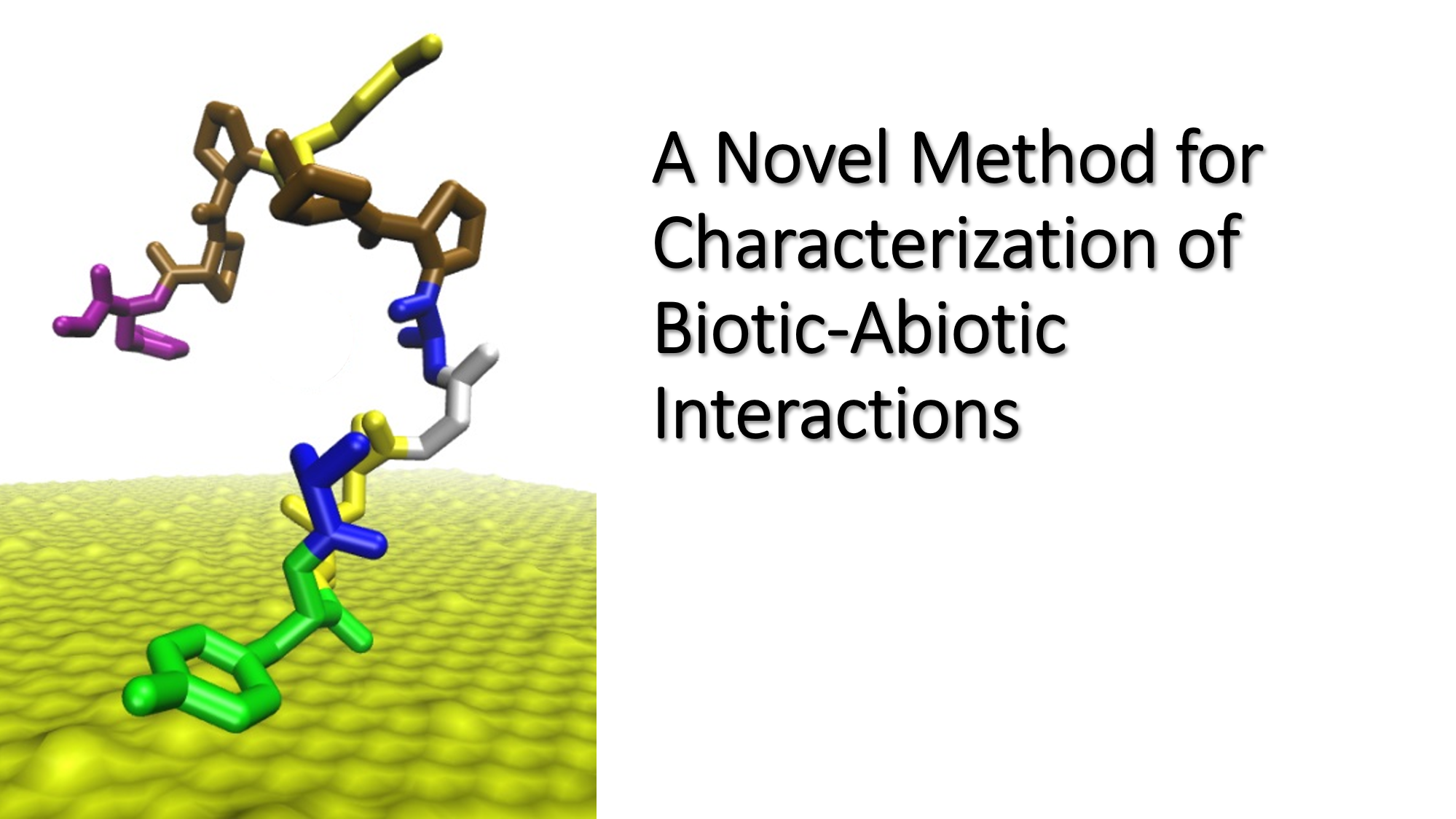 We're excited to share our Biological & Nanoscale Technologies team's latest publication! This work is titled "Gamma estimator of Jarzynski equality for recovering binding energies from noisy dynamic data sets" and is published in Nature Communications. UES scientists Dr. Zhifeng Kuang and Kristi M. Singh collaborated with scientists from Nottingham Trent University and the Air Force Research Laboratory in this work.
We're excited to share our Biological & Nanoscale Technologies team's latest publication! This work is titled "Gamma estimator of Jarzynski equality for recovering binding energies from noisy dynamic data sets" and is published in Nature Communications. UES scientists Dr. Zhifeng Kuang and Kristi M. Singh collaborated with scientists from Nottingham Trent University and the Air Force Research Laboratory in this work.
Read also: Recent Biological & Nanoscale Technologies Research
First, a little background. In 2019, American Physical Society awarded UMD Professor Christopher Jarzynski the Lars Onsager Prize for his discovery of the Jarzynski equality (JE). It relates non-equilibrium fluctuations to equilibrium free energy differences. In his seminal paper, he thought the JE's applications were limited to slow processes with fluctuations not much greater than kT. This condition pretty much ruled out biological systems of interest.
UES scientist Dr. Zhifeng Kuang wanted to find a way to extend JE to those biological applications. In this paper, Dr. Kuang proposed a gamma estimator of JE to allow fluctuations to be greater than 100 kT. "The gamma estimator has shown to be applicable to single-molecule atomic force measurements of peptide adsorption on surface. Implementing the gamma estimator produced the first publicly available code for reconstructing absorption free energy from noisy single-molecule atomic force data."
Another UES scientist, Kristi M. Singh, performed the atomic force microscopy (AFM) experiments. Dr. Kuang used that data to process and extract the binding free energy of single molecule peptide-surface interactions. "By carefully controlling the AFM tip modification approach and experimental settings, we are able to generate force-distance curves showing the interaction forces as a peptide is pulled away from a surface. This data can then be used to calculate the absorption free energy. Since we generate thousands of these curves, evaluating them all manually is very time consuming. Dr. Kuang developed the code for both identifying the AFM curves that show peptide binding as well as calculating the free energy from the selected curves."
Dr. Kuang goes on to discuss how this work opens the door for future applications. "This new method in extracting the binding free energy for understanding biotic-abiotic interactions is useful in biomaterials discovery and design for sensing, antifouling, and biomedical applications."
You can read the full article online in Nature Communications and access the code here.
Read also: Aircraft Decontamination Team Fights COVID-19
Questions? Contact us here. Connect with us on Facebook, Twitter, LinkedIn, and Instagram.
